library(titanic)
library(tidyverse)
titanic_df <- titanic_train %>%
as_tibble() %>%
janitor::clean_names() %>%
mutate(
survived = case_when(
survived == "1" ~ "survived",
TRUE ~ "died"
) %>%
as.factor() %>%
fct_relevel(c("died", "survived"))
) %>%
mutate(
cabin = case_when(
cabin == "" ~ NA_character_,
TRUE ~ cabin
),
embarked = case_when(
embarked == "" ~ NA_character_,
TRUE ~ embarked
),
across(c(pclass, sex, embarked), as.factor),
passenger_id = as.character(passenger_id)
)
library(tidymodels)
set.seed(123)
titanic_split <- initial_split(titanic_df, 0.75, strata = survived)
titanic_training <- titanic_split %>% training()
titanic_testing <- titanic_split %>% testing()
logistic_model <- logistic_reg(penalty = 0.1, mixture = 0) %>%
set_engine("glmnet") %>%
set_mode("classification")
titanic_recipe <- recipe(
survived ~ fare + sex + sib_sp + parch + pclass,
data = titanic_training
) %>%
step_corr(all_numeric(), threshold = 0.8) %>%
step_normalize(all_numeric()) %>%
step_dummy(all_nominal(), -all_outcomes(), one_hot = TRUE)
titanic_recipe_prep <- titanic_recipe %>% prep(titanic_training)
titanic_training_prep <- titanic_recipe_prep %>% bake(titanic_training)
titanic_testing_prep <- titanic_recipe_prep %>% bake(titanic_testing)
logistic_fit <- logistic_model %>%
fit(survived ~ ., titanic_training_prep)
levels(titanic_training_prep$survived)
## [1] "died" "survived"
tidy(logistic_fit)
#
#
#
#
#
#
#
#
#
#
#
#
pred_class <- logistic_fit %>%
predict(titanic_testing_prep, type = "class")
pred_prob <- logistic_fit %>%
predict(titanic_testing_prep, type = "prob")
titantic_testing_results <- titanic_testing %>%
select(survived) %>%
bind_cols(pred_class, pred_prob)
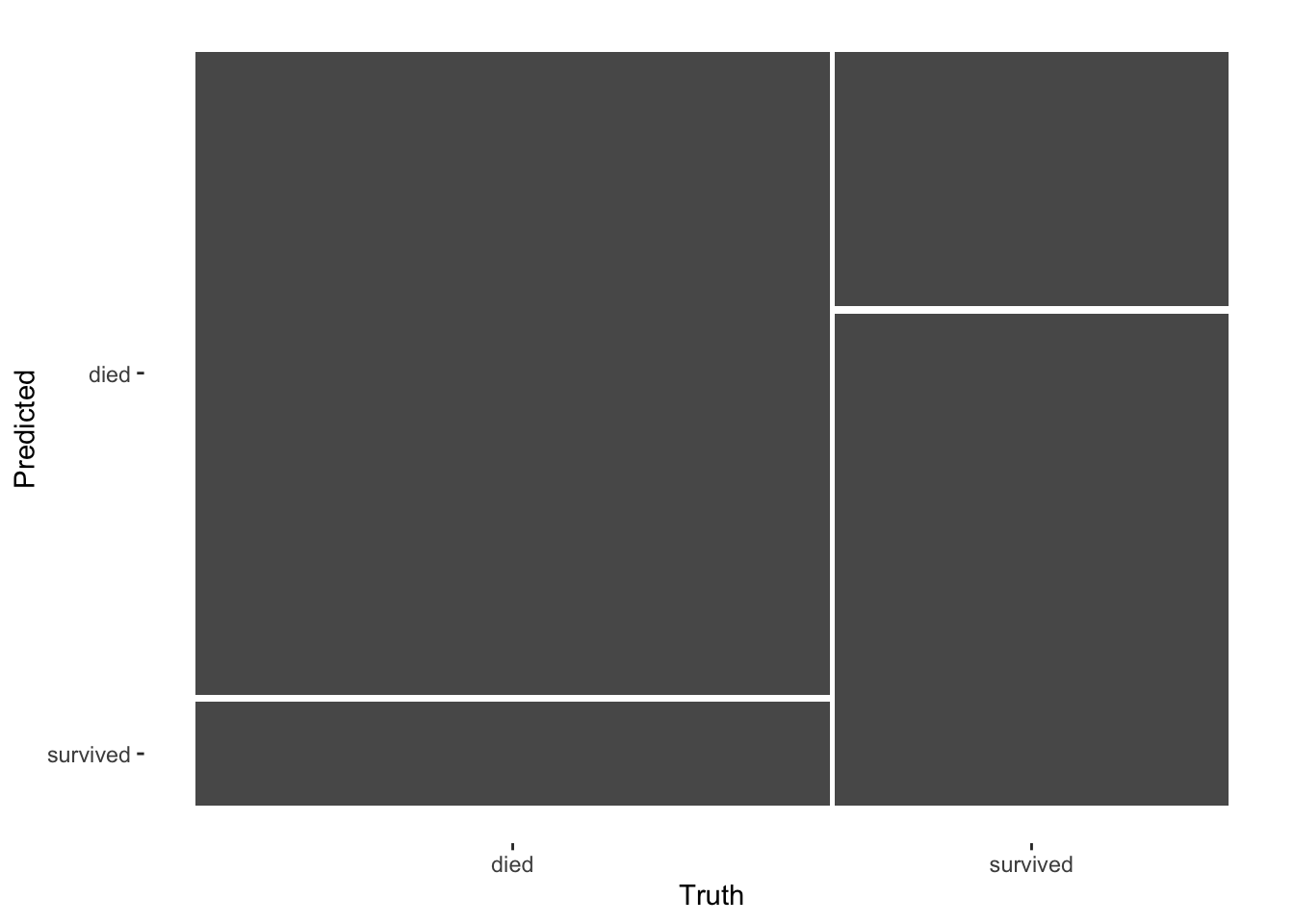
titantic_testing_results %>%
conf_mat(survived, .pred_class) %>%
autoplot(type = "mosaic")
titanic_metrics <- metric_set(roc_auc, sens, spec, accuracy)
titantic_testing_results %>%
titanic_metrics(
truth = survived, estimate = .pred_class, .pred_survived,
event_level = "second"
)
#
#
#
#
#
#
#

